-
Notifications
You must be signed in to change notification settings - Fork 2
Simulating
Aim: Simulate a tree
Ingredients
- Initial random tree
- Simulation parameters
Problem
In this case example we want to simulate the evolution of a tree through time. We can do that using a simple for loop and randomly adding new tips using the addTip() function. To make the demonstration more interesting we've biased the placement and removal of tips based on their evolutionary distinctness calculated using the calcPrtFrPrp() function. By biasing with evolutionary distinctness, we can generate phylogenetic trees that are very similar in shape to empirically derived trees. See Bennett et al. (2017)
Process and code
First let's read in a simple balanced tree.
tree_string <- "((A:1.0,B:1.0):1.0,(C:1.0,D:1.0):1.0);"
tree_age <- 2
tree <- readTree(text=tree_string)And then set some parameters. Let's run the simulation for 200 iterations, with an initial burnin birth rate of 3 followed by a birth rate of 1. The death rate can be 1.
iterations <- 200
burnin <- 10
d <- 1
b <- 3 # birth for burnin
b_true <- 1 # birth after burninNow for the simulation loop. In each iteration, we need to calculate the evolutionary distinctness of each tip in the tree. To do this we can use the calcPrtFrPrp() which allows us to calculated evolutionary distinctness for a subset of tips, i.e. those ones that are extant, extnt. Secondly, we can then decide whether we want to add or remove a tip in this time step. If adding, we create a new tip ID and add a tip using the addTip() function. If removing, we can just add the removed tip to the extnct vector. Whenever adding or removing, we are biasing the selection of tips by their evolutionary distinctness. In the final part of the iteration, we update the branch lengths of all extant tips by adding 1 to them.
extnt <- tree["tips"]
extnct <- NULL
for(i in 1:iterations) {
if(length(extnt) < 3) {
stop('Too few extant tips remaining!\nRun again or increase birth before burnin.')
}
if(i > burnin) {
b <- b_true
}
cat('.... i=[', i, ']\n', sep='')
# calculate fair proportion
fps <- calcPrtFrPrp(tree, tids=extnt, ignr=extnct)
# add/remove based on b and d
to_add <- sample(c(TRUE, FALSE), size=1, prob=c(b,d))
if(to_add) {
sid <- sample(extnt, prob=1/fps, size=1) # sister ID of the new tip
tid <- paste0('t', i) # new tip ID
tree <- addTip(tree, tid=tid, sid=sid, strt_age=0, end_age=0,
tree_age=tree_age)
extnt <- c(extnt, tid)
} else {
tid <- sample(extnt, prob=1/fps, size=1)
extnct <- c(extnct, tid)
extnt <- extnt[extnt != tid]
}
# grow tree
spns <- getNdsSlt(tree, slt_nm="spn", ids=extnt)
tree <- setNdsSpn(tree, ids=extnt, vals=spns+1)
tree_age <- tree_age + 1
}Running our simulation may lead to the tree becoming too small (< 3 tips). We've added an if statement preventing the simulation continuing if this is the case. We can avoid this issue by increasing the births before burnin.
After we have simulated our tree, we can take a peek using APE's plot function. We'll look at the full tree showing the simulation history and the final reconstructed phylogeny of all extant tips.
extnt_tree <- rmTips(tree, tids=getDcsd(tree))
par(mfrow=c(1,2))
plot(as(tree, 'phylo'), show.tip.label=FALSE, main='Fossil record')
plot(as(extnt_tree, 'phylo'), main='Reconstructed')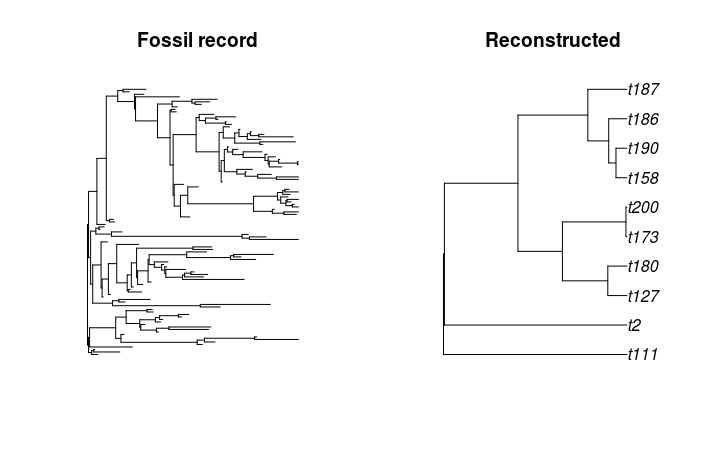
Next page: Measuring community turnover