-
Notifications
You must be signed in to change notification settings - Fork 12
Description
So I got a bit more info on our data originally described here #36 . In our case, our "adapter" sequence actually consists of several parts that are concatenated together in the following order:
Tn5 transposase sequence (same across all cells)+cell-specific barcode sequence (different for each cell)+adapter sequence (in the conventional sense; same across all cells)
This is why our so-called "adapters" were different for each cell. So I tried an alternative strategy so that we could have just one adapter per sample, while keeping different barcodes per cell. I did this by splitting this adapter into two parts
new "barcodes" = part 1 + part 2: Many sequences (1 per cell) to be used in theultraplex -bargument.new "adapter" = part 3: One unique sequence to be used in theultraplex -aargument (along with its reverse complement as the-a2argument).
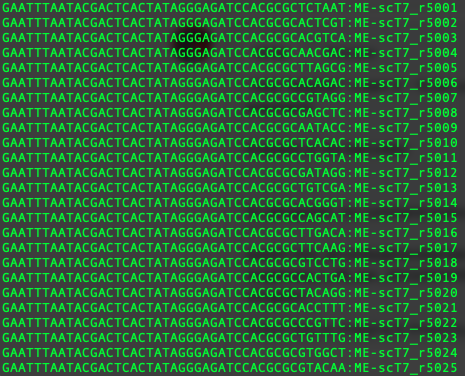
Example of the barcodes:
So altogether, my command looks like:
export root_dir=/rds/general/project/neurogenomics-lab/live/Data/tip_seq/raw_data/scTIP-seq/phase_1_06_apr_2022/X204SC21103786-Z01-F003/raw_data/scTS_1
ultraplex \
-i $root_dir/scTS_1_EKDL220003595-1a_HHN53DSX3_L1_1.fq.gz \
-i2 $root_dir/scTS_1_EKDL220003595-1a_HHN53DSX3_L1_2.fq.gz \
-b $root_dir/barcodes.csv \
-d $root_dir/demultiplexed/ \
-a TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG \
-a2 CTGTCTCTTATACACATCTGACGCTGCCGACGA \
-t 38
However, for some reason running ultraplex this way leads to an explosion of memory usage! On an HPC job submission with 128Gb of RAM / 40 cores, ultraplex consistently exceeds the memory limit thus killing the job. Adjusting the number of threads in -t from 38 to 20 to 8 to 4 didn't seem to help this issue and the jobs were killed each time.
To make sure this wasn't an HPC-specific problem, I also ran the same command on our lab's 250+Gb RAM / 64-core Linux machine. However even then ultraplex quickly uses up all available memory (I stopped the process before it could crash the machine).
This seems bizarre to me since ultraplex was very efficient when I previously ran it on these same fastq files using 6-basepair-long barcodes and without specifying the adapter sequences via flags. But I'm not quite sure which aspect of how I'm running ultraplex is causing this explosion in memory usage.
Let me know if I'm doing something terribly wrong here!
Many thanks,
Brian
Conda env versions
Note: I'm using the latest conda distribution of ultraplex, not the devel version described here which I haven't quite figured out how to run just yet.
Details
# packages in environment at /home/bms20/anaconda3/envs/ultra:
#
# Name Version Build Channel
_libgcc_mutex 0.1 conda_forge conda-forge
_openmp_mutex 4.5 1_gnu conda-forge
ca-certificates 2021.1.19 h06a4308_0
certifi 2020.12.5 py36h5fab9bb_1 conda-forge
dataclasses 0.7 pyhe4b4509_6 conda-forge
dnaio 0.5.0 py36h4c5857e_0 bioconda
isa-l 2.30.0 ha770c72_2 conda-forge
ld_impl_linux-64 2.35.1 hea4e1c9_2 conda-forge
libffi 3.3 h58526e2_2 conda-forge
libgcc-ng 9.3.0 h2828fa1_18 conda-forge
libgomp 9.3.0 h2828fa1_18 conda-forge
libstdcxx-ng 9.3.0 h6de172a_18 conda-forge
ncurses 6.2 h58526e2_4 conda-forge
openssl 1.1.1j h7f98852_0 conda-forge
pigz 2.5 h27826a3_0 conda-forge
pip 21.0.1 pyhd8ed1ab_0 conda-forge
python 3.6.13 hffdb5ce_0_cpython conda-forge
python-isal 0.5.0 py36h8f6f2f9_0 conda-forge
python_abi 3.6 1_cp36m conda-forge
readline 8.1 h27cfd23_0
setuptools 52.0.0 py36h06a4308_0
sqlite 3.34.0 h74cdb3f_0 conda-forge
tk 8.6.10 h21135ba_1 conda-forge
ultraplex 1.1.3 py36h4c5857e_0 bioconda
wheel 0.36.2 pyhd3deb0d_0 conda-forge
xopen 1.1.0 py36h5fab9bb_1 conda-forge
xz 5.2.5 h516909a_1 conda-forge
zlib 1.2.11 h516909a_1010 conda-forge